A single-cell multi-omic and spatial atlas of B-cell lymphomas reveals differentiation drives intratumor heterogeneity
September 27, 2024·,,,,,,,,,,,,,,, ,,,,,,,·
0 min read
,,,,,,,·
0 min read
Donnacha Fitzgerald
Tobias Roider
Marc-Andrea Baertsch
Artur Kibler
Anastasiia Horlova
Erin Chung
Harald Vöhringer
Anna Mathioudaki
Bettina Budeus
Verena Passerini
Johannes Mammen
Linsha Li
Léandra Caillé
Felix Czernilofsky
Peter-Martin Bruch
Nora Liebers
Matthias Meyer-Bender
Reiner Siebert
Oliver Weigert
Judith Zaugg
Garry Nolan
Marc Seifert
Sascha Dietrich
Wolfgang Huber
Abstract
Intratumor heterogeneity underpins cancer pathogenesis and evolution, although it is typically considered independent from the differentiation processes that drive physiological cell-type diversity. As cancer types and subtypes arise from different cell types, we investigated whether cellular differentiation influences intratumor heterogeneity.
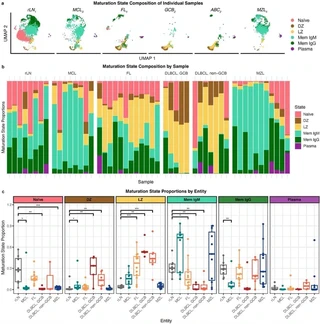
Nodal B-cell non-Hodgkin lymphomas are a diverse set of cancers originating from different stages of B-cell maturation. Through single-cell transcriptome and surface epitope profiling (CITE-Seq) of diffuse large B-cell, mantle cell, follicular, and marginal zone lymphomas in addition to reactive lymph nodes from 51 patients, we found multiple B-cell maturation states within tumors. Intratumor maturation states emerged from the same clone, revealing divergent differentiation from a shared cell of origin. Maturation state composition varied across subtypes and tumors, which encompassed mixed cell-of-origin diagnostic subtypes.
Through highly multiplexed immunohistochemistry (CODEX) of samples from 19 of these patients, we found that intratumor maturation states inhabited distinct spatial niches, displaying cellular interactions and regulatory networks typical of their maturation states while harboring different genetic variants. By deconvoluting intratumor maturation states from a microarray dataset of 507 patients, we identified risk groups within diagnoses with striking differences in survival, including IgM memory-enriched germinal center B-cell (M = 1.9 vs >10 years, p = 0.00039) and activated B-cell (M = 2.4 vs 9.6 years, p = 0.016) diffuse large B-cell lymphoma, and dark zone-enriched follicular lymphoma (M = 8.6 vs 13 years; p = 0.0019). Our findings reveal cellular differentiation remains plastic in B-cell lymphomas, driving tumor variation, evolution, and response.
Nodal B-cell non-Hodgkin lymphomas are a diverse set of cancers originating from different stages of B-cell maturation. Through single-cell transcriptome and surface epitope profiling (CITE-Seq) of diffuse large B-cell, mantle cell, follicular, and marginal zone lymphomas in addition to reactive lymph nodes from 51 patients, we found multiple B-cell maturation states within tumors. Intratumor maturation states emerged from the same clone, revealing divergent differentiation from a shared cell of origin. Maturation state composition varied across subtypes and tumors, which encompassed mixed cell-of-origin diagnostic subtypes.
Through highly multiplexed immunohistochemistry (CODEX) of samples from 19 of these patients, we found that intratumor maturation states inhabited distinct spatial niches, displaying cellular interactions and regulatory networks typical of their maturation states while harboring different genetic variants. By deconvoluting intratumor maturation states from a microarray dataset of 507 patients, we identified risk groups within diagnoses with striking differences in survival, including IgM memory-enriched germinal center B-cell (M = 1.9 vs >10 years, p = 0.00039) and activated B-cell (M = 2.4 vs 9.6 years, p = 0.016) diffuse large B-cell lymphoma, and dark zone-enriched follicular lymphoma (M = 8.6 vs 13 years; p = 0.0019). Our findings reveal cellular differentiation remains plastic in B-cell lymphomas, driving tumor variation, evolution, and response.
Type
Publication
bioRxiv
Authors
Authors
Authors
Authors
Authors
Authors
Authors
Authors
Authors
Authors
Authors
Authors
Authors
Authors
Authors
Authors

Authors
Matthias Meyer-Bender
(he/him)
Bioinformatician
I am a PhD candidate at the European Molecular Biology Laboratory (EMBL) in the group of Wolfgang Huber. My current work focuses on developing open-source software for the processing and analysis of large-scale spatial omics datasets (spatialproteomics). I am passionate about applying computational methods, including machine learning and AI, to answer questions of biomedical relevance. Previously, I studied bioinformatics at the Technical University of Munich (TUM) and Ludwig Maximilian University of Munich (LMU). I also contribute to open-source projects such as scverse.
Authors
Authors
Authors
Authors
Authors
Authors
Authors